
Engineering
Design
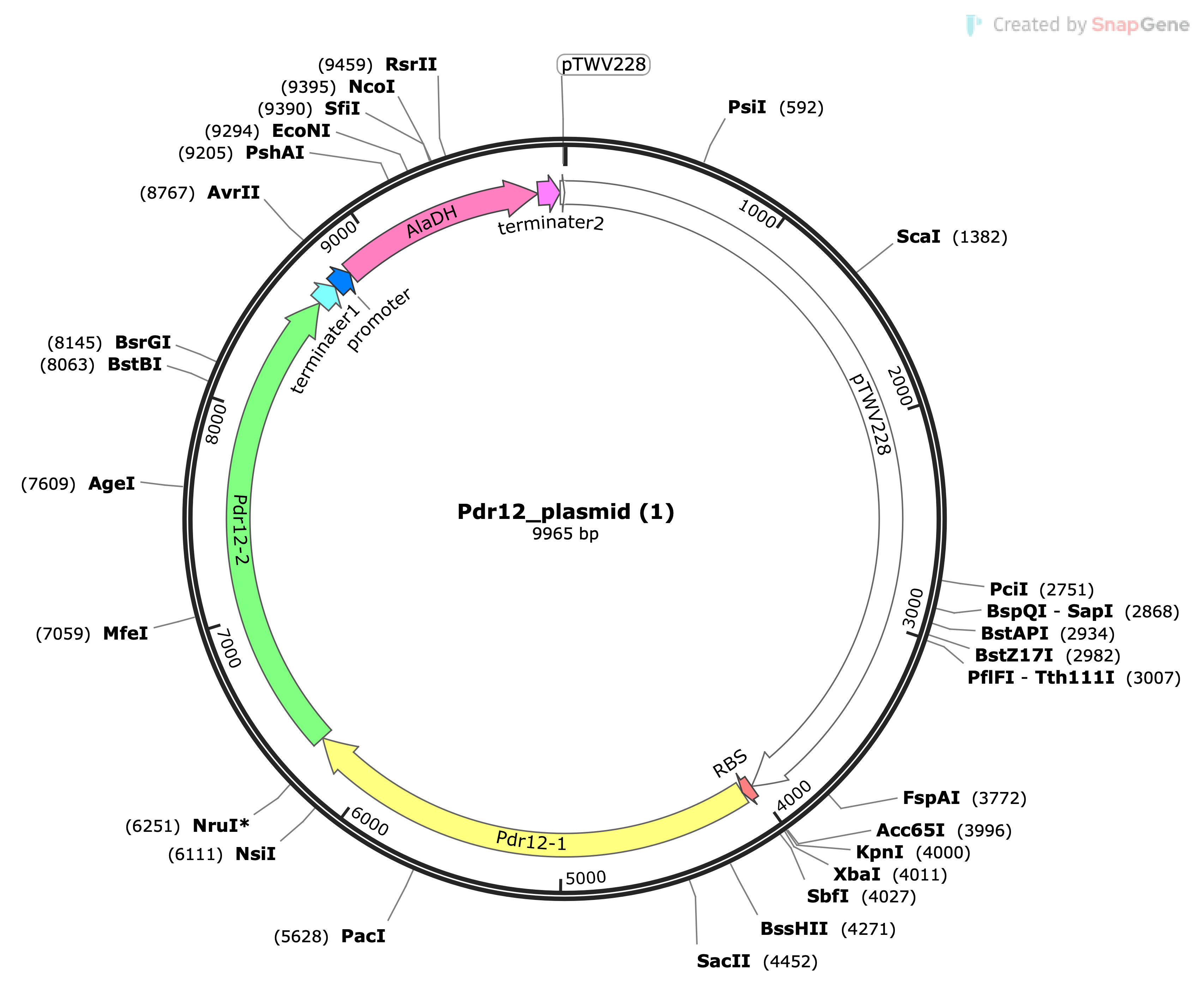
In the design stage of your experiment, the objective was to enhance E. coli's metabolic pathway for converting nylon-6 oligomers into pyruvic acid. This modified biological system in a bacterial chassis (E. coli) includes the AlaDH gene for amino transfer from alanine to pyruvate, the Pdr12 gene for pyruvate transport, and the pTWV228 plasmid for cloning. These components were assembled using Gibson Assembly. The design’s efficacy relies on the interaction of these biological parts, whose functions are well-characterized in scientific literature. The performance of this system can be modeled computationally, and testing involves measuring the conversion rate of alanine to pyruvate in the modified E. coli. Successful functioning would be indicated by a higher conversion rate compared to unmodified strains.
Build
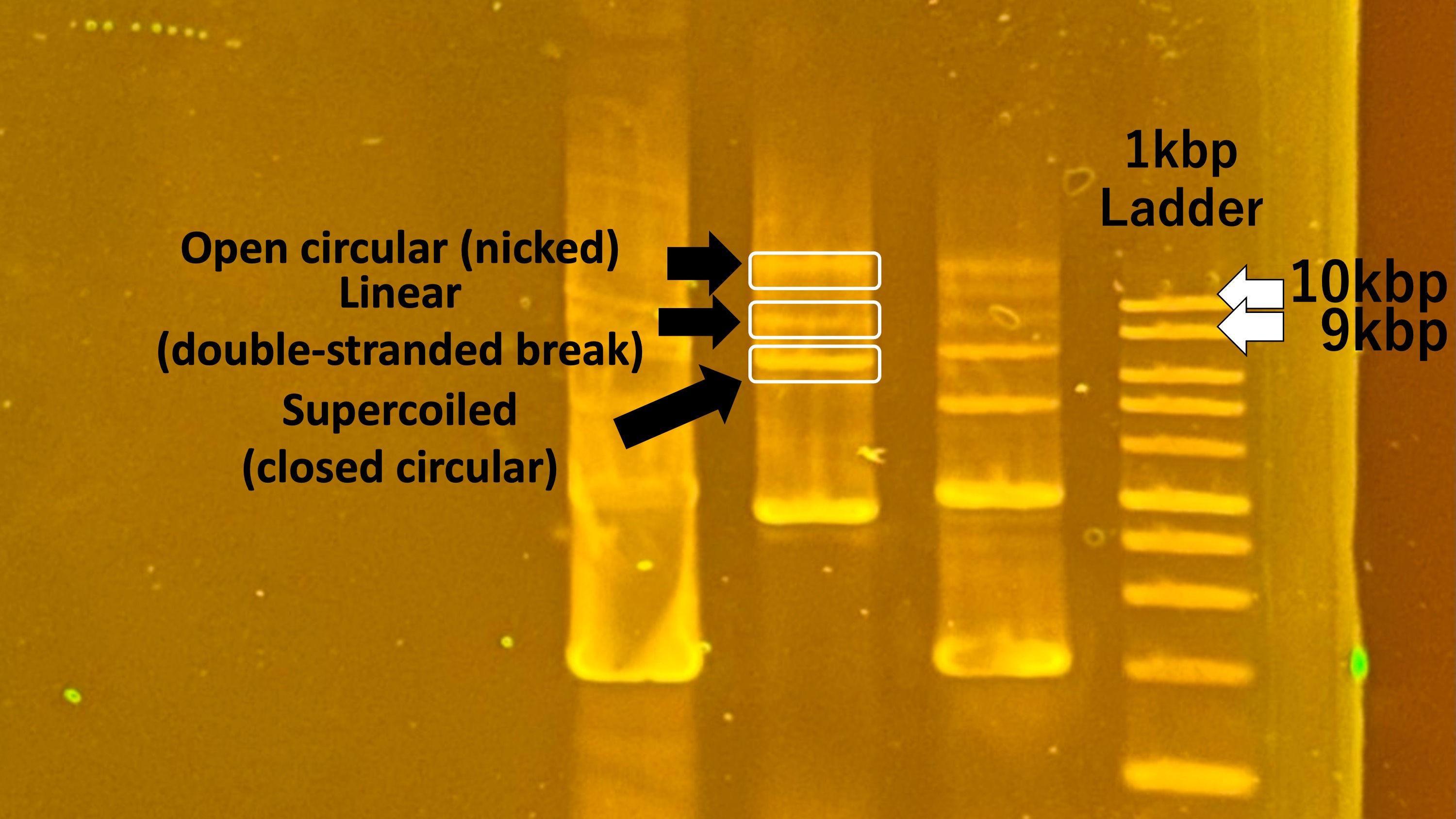
In the build stage of your experiment, the system was created as a cohesive unit by integrating the AlaDH gene, Pdr12 gene, and pTWV228 plasmid into the E. coli chassis. This was achieved using Gibson Assembly, which can create scar sites at DNA junctions. The assembled vector was then introduced into the E. coli using the heat shock method. Screening for correct builds involved colony selection and the use of antibiotic resistance markers for verification.
Test
In the test stage of our experiment, the functionality of the genes (AlaDH and Pdr12) in the designed plasmid was evaluated. This was done by culturing the transformed E. coli in a medium containing an alanine solution and then measuring the concentration of pyruvic acid. This culturing commenced only after the optical density (OD) in LB medium exceeded 2.0. The pyruvic acid levels were determined using a specialized kit, measuring absorbance at 575 nm. The data obtained in terms of absorbance was used to calculate the pyruvic acid concentration, based on the relationship between absorbance and concentration established from blank tests. Additionally, a comparison was made by inducing the expression of the Pdr12 gene with IPTG, examining any differences in pyruvic acid production.

Learn
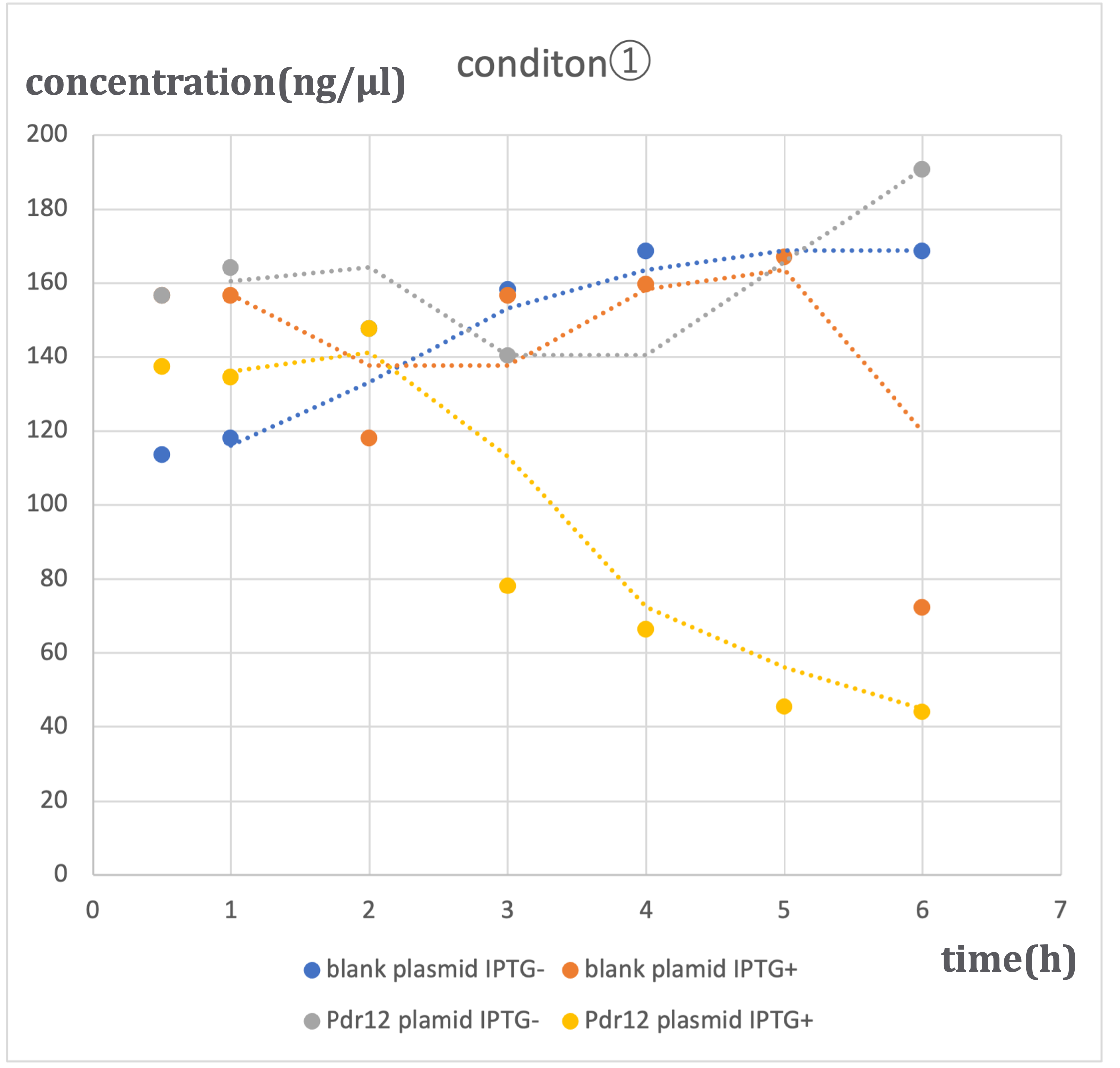
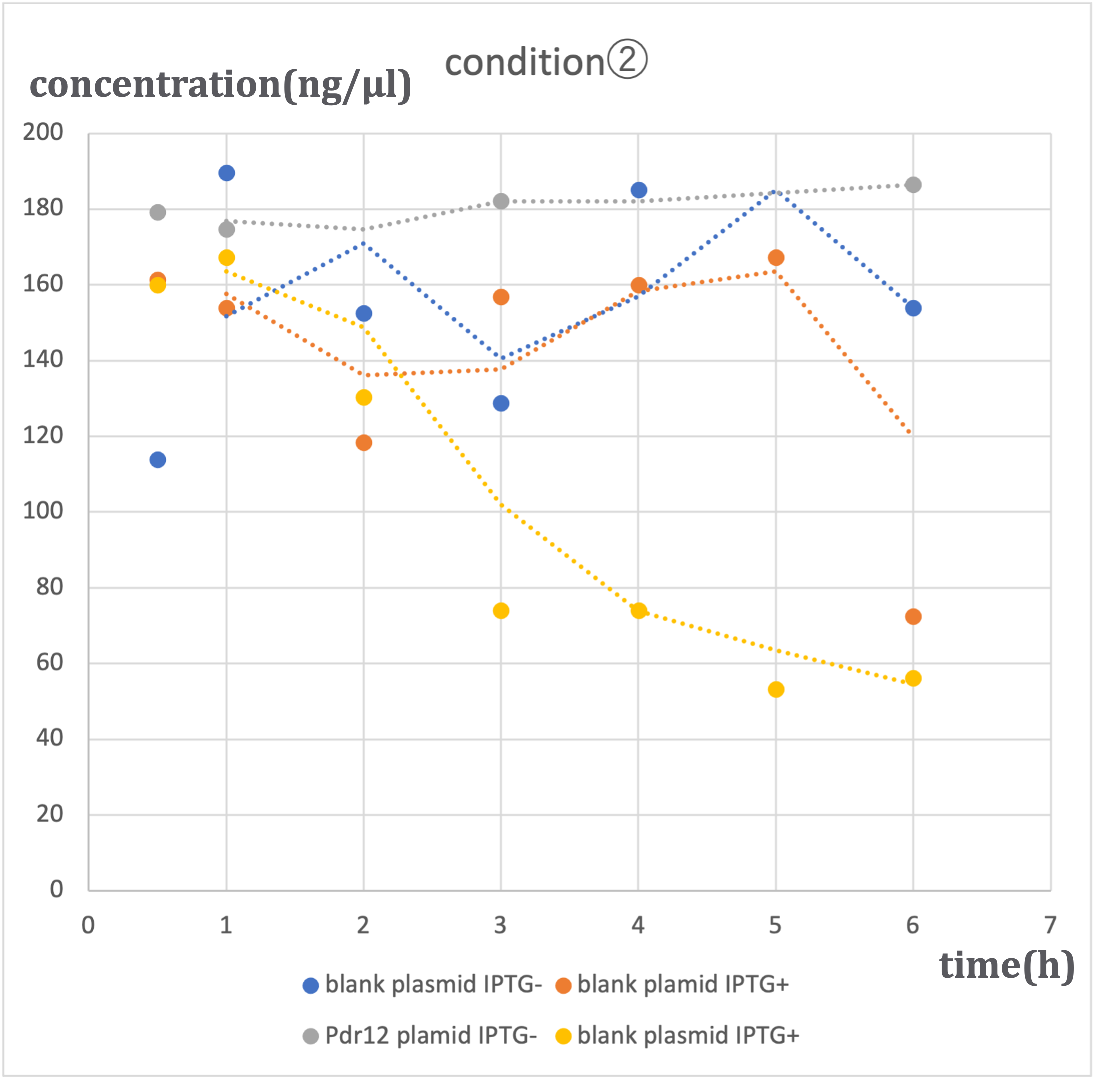
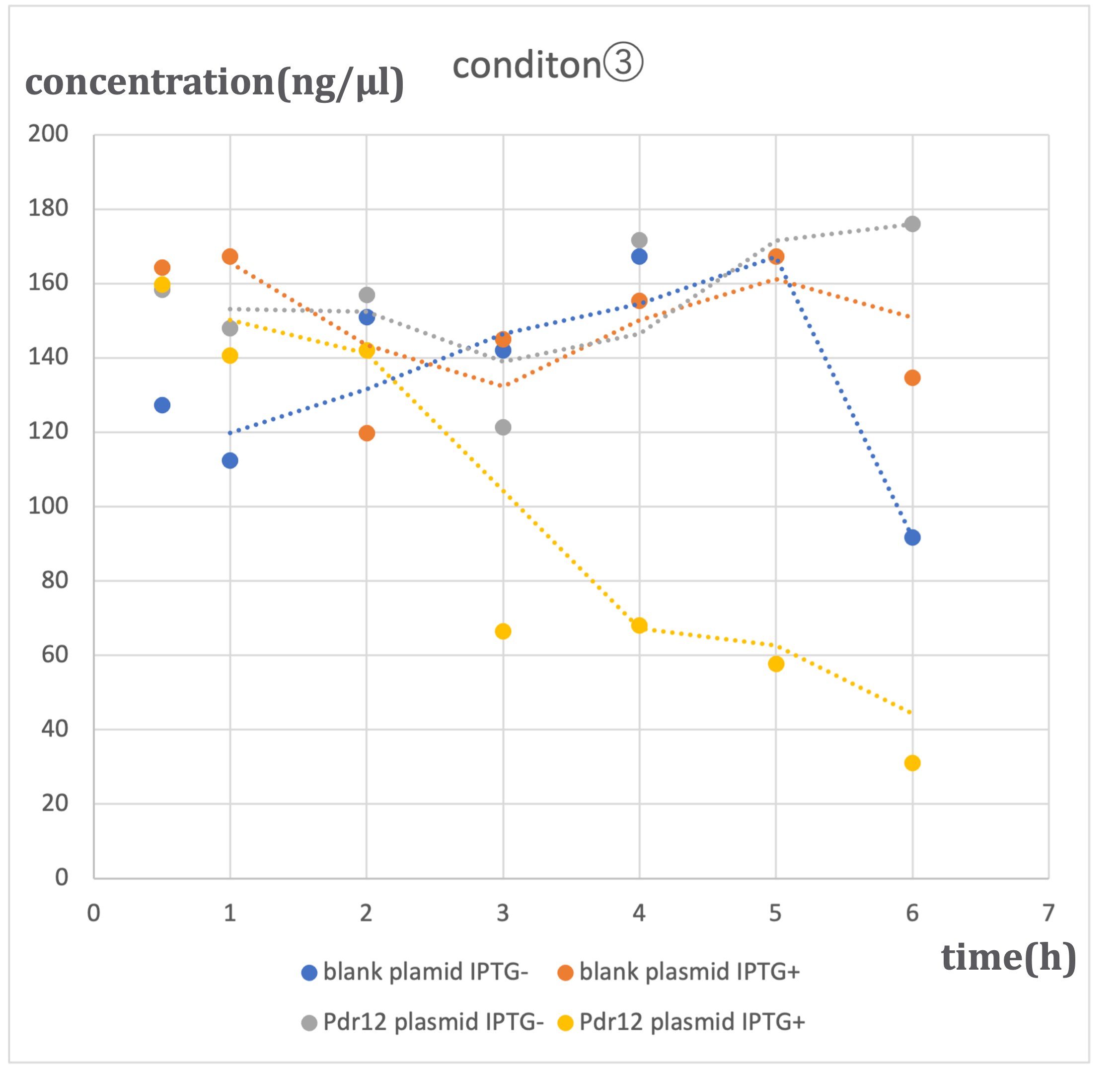
Under three different conditions with varying alanine concentrations (500 μM - ①, 1 mM - ②, 2 mM - ③), similar trends were observed. In conditions where blank plasmid was induced with IPTG (hereinafter referred to as 'blank plasmid IPTG+'), as well as conditions without IPTG induction (hereinafter referred to as 'blank plasmid IPTG-'), and conditions where Pdr12 plasmid was not induced with IPTG (hereinafter referred to as 'Pdr12 plasmid IPTG-'), a slight increase in pyruvic acid concentration was observed over time. In contrast, in conditions where Pdr12 plasmid was induced with IPTG (hereinafter referred to as 'Pdr12 plasmid IPTG+'), a decrease in pyruvic acid concentration was observed at all alanine concentrations.
In the 'Pdr12 plasmid IPTG+' conditions, an increase in pyruvic acid concentration was expected, but the actual results differed. This result suggests that IPTG induction led to an increased expression of the Pdr12 protein, causing an increase in the pyruvic acid demand within Escherichia coli, which was transported into the bacteria through membrane proteins. Despite the knockout of the aceF gene, the effect on pyruvic acid consumption was limited, and the fundamental E. coli demand remained unchanged.
In the 'blank plasmid IPTG+' conditions, a significant decrease in pyruvic acid concentration was not observed, implying that IPTG induction alone, which induces the expression of β-galactosidase, does not lead to a significant change in pyruvic acid demand. On the other hand, as the Pdr12 protein is expressed from a gene of approximately 4000 bp, it has the potential to have a substantial impact on E. coli metabolism.
As a result, the system did not function as expected. The Pdr12 protein was originally found in yeast and is challenging to function properly within E. coli due to its membrane protein nature. In the future, we plan to confirm whether this protein is expressed at all using SDS-PAGE and visualize how pyruvic acid is moving within E. coli in real-time using an approach that marks pyruvic acid.
References
- Casal, Margarida, et al. "Transport of carboxylic acids in yeasts." FEMS microbiology reviews 32.6 (2008): 974-994.
- Hazelwood, Lucie A., et al. "A new physiological role for Pdr12p in Saccharomyces cerevisiae: export of aromatic and branched-chain organic acids produced in amino acid catabolism." FEMS yeast research 6.6 (2006): 937-945.
- Liu, Ke, et al. "Biotransformation and chiral resolution of d, l‐alanine into pyruvate and d‐alanine with a whole‐cell biocatalyst expressing l‐amino acid deaminase." Biotechnology and Applied Biochemistry 67.4 (2020): 668-676.
- Hossain, Gazi Sakir, et al. "Transporter engineering and enzyme evolution for pyruvate production from D/L-alanine with a whole-cell biocatalyst expressing L-amino acid deaminase from Proteus mirabilis." RSC advances 6.86 (2016): 82676-82684.
- Tomar, A., M. A. Eiteman, and E. Altman. "The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli." Applied microbiology and biotechnology 62 (2003): 76-82.
- Dave, Ushmaben Chandrakantbhai, and Ravi-Kumar Kadeppagari. "Alanine dehydrogenase and its applications–A review." Critical reviews in biotechnology 39.5 (2019): 648-664.
- Vieira J and Messing J.Methods in Enzymology. (1987) 153: 3-11.
- Boliver F, Rodrigues R L, Green, P J, Betlach M C, Heynecker H L,Boyer H W, Grosa J H, and Falkow S.Gene. (1977) 2: 95-113.