Engineering
Our aim was to use the BioBrick format for the plasmids pRS426 and YIplac204 as well as the inserts, so as to produce phytase enzyme in the host organism, which we chose to be Saccharomyces cerevisiae.
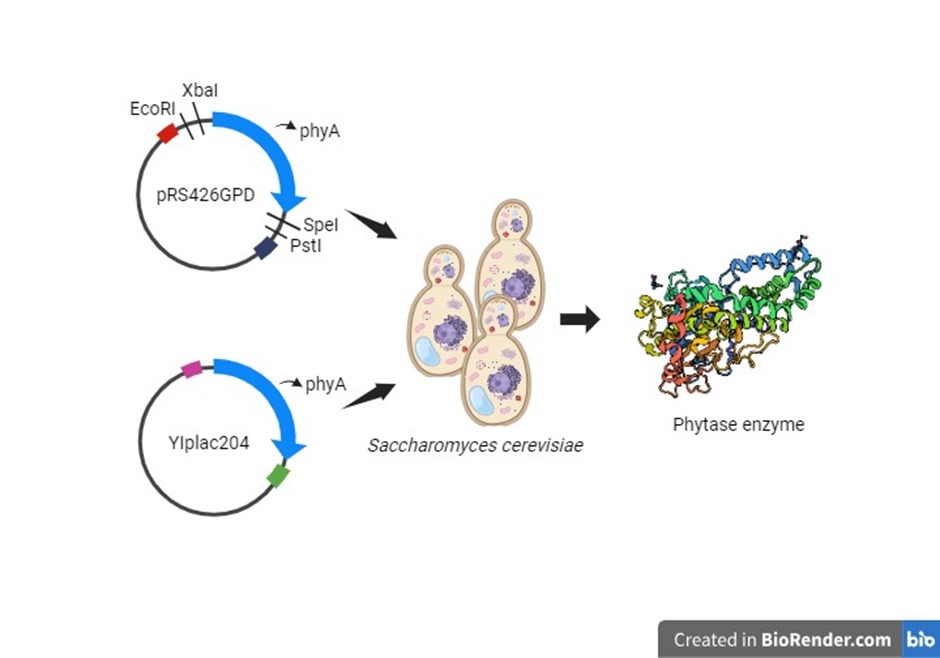
Amplification of the genes

1
Design: Amplifying the genes using a high-fidelity polymerase in a normal PCR reaction so that we could further perform restriction-digestion and then ligate them to our plasmid (pRS426GPD)
Build: Set up the PCR reaction
Test: Gel electrophoresis showed bands of much smaller length than that of the gene of interest.
Learn: PCR reaction was not successful.

2
Design: Since the high-fidelity enzyme (phusion) did not work, we decided to use Q5 and a combination of Taq and Pfu polymerase in varying proportions for the amplification.
Build: Set up the PCR reaction
Test: Gel electrophoresis did not show bands indicating desired length.
Learn: None of the afore mentioned polymerases worked. PCR was unsuccessful.
After multiple tries at trouble-shooting for 3 weeks, we arrived at the conclusion that the polymerases we were using were degrading our primers very quickly because they had longer “tails”

1
Design: We then decided on using a long-range polymerase for amplification of the genes.
Build: Set up the PCR
Test: Gel electrophoresis of the purified PCR product using a long-range polymerase showed bands of desirable lengths.
. . . . . . . . . . . . . . . . . . . . . . . . . . . . Learn: A long-range polymerase showed positive results for our PCR

Amplification of the plasmid fragments and joining the fragments using SOE PCR

1
Design: Since we had to design primers such that we could avoid the PstI site in our plasmid, we decided to amplify the individual fragments and then use splice over extension (SOE) PCR to join the fragments together. We will also be requiring an equimolar mixture of the fragments for performing SOE PCR.
Build: Set up the PCR reaction for amplification; quantify the fragment mixtures; set up SOE PCR.
Test: Bands at desirable lengths obtained for the amplification; No bands visible after the setting up of SOE PCR.
Learn: SOE PCR was not successful and needed to be troubleshooted.
After multiple changes in our protocol, our SOE PCR was still unsuccessful! To show proof of concept, we decided to use different restriction sites for the amplification of the genes to introduce into the plasmid.
Restriction-digestion (RD) of the inserts and the plasmid

1
Design:Producing sticky ends using REases to facilitate ligation of the fragments.
Build: Set up RD followed by gel purification
Test: Gel electrophoresis showed bands at desirable length
Learn: RD was successful
Ligation was set up for these fragments and E. coli transformation was carried out:
Image Credits:Team ICT-Mumbai 2022
Adding the recombinant plasmid into yeast cells

1
Design:Isolate the recombinant plasmid from the transformed E. coli and insert into Saccharomyces cerevisiae
Build: Yeast transformation protocol was carried out and the culture was plated on medium – agar + YNB + complete synthetic medium w/o URA + glucose
Test: Colonies were observed on the second day after plating. Observing under the microscope confirmed it was yeast.
Learn: Yeast transformation was successful
Phytate content estimation

1
Design:We decided to estimate the decrease in phytate content in a given sample by measuring one of the products formed after the breakdown of phytic acid upon the addition of our enzyme after intervals of half hour and 1h. We decided to use the Fiske-Subarrow method for the estimation of free inorganic phosphate in the reaction mixture after those stipulated time intervals.
Build: Carried out the protocol for the Fiske-Subarrow method, a colorimetric method for the estimation of inorganic phosphate.
Test: While preparation of the samples for the estimation, we observed our blank was also showing colour upon the addition of the reagent.
. . . . . . . . . . . . . . . . . . . . .Learn: Phytic acid also gives positive results for the Fiske-Subbarrow method. An alternate method for the estimation of phytate or modifications to the existing method required.