Lichenysin gene cluster
Preparation of vectors:
Plasmid, pSIM7 was isolated from its culture. The vector was digested with SmaI and BglII, having a fallout of about 1816 bp, after which the linearized band was cut and gel extraction was performed with it. Gel extraction of the linear band was performed and then eluted. All of the gels were run on 1% agarose.

Preparation of insert: As the length of the insert to be incorporated was 26.6 Kb, special primers with overhangs were designed so that specific binding can take place. Unfortunately, even after multiple attempts of optimizing temperature, template concentrations with both colony PCR and PCR of isolated genomic DNA of B. licheniformis an amplicon could not be obtained.

Dual Promoter System
Preparation of inserts:
We revived the synthesized genes for alasan, and rhlAB, and made working stocks of 100ng/ul. We also revived their primers and made working stocks of those to use in our PCR reactions. The gene for Ptet was obtained by amplification of the 156bp fragment from a plasmid carrying the promoter. We obtained amplification of all the three gene fragments we set out to ultimately clone, Ptet, aln, and rhlAB with Taq polymerase and using the respective annealing temperatures as was found out from the NEB Tm calculator, 52°C, 51°C and 62°C respectively with extension times as was dictated by the speed of the Taq polymerase we used as well as the gene length. For insert digestion we then digested our gel extracted PCR products with the sets of enzymes we needed to clone them with. The gene for alasan, aln was digested with NdeI and HindII, Ptet with BglII and SphI, and rhlAB was digested with XbaI and SphI. The reactions were digested for 8 hours to 12 hours depending on the concentration of the gel extracted elute. They were then PCR purified and ran on 1% agarose gel. The images have been added below:


Preparation of vectors: Plasmids, pTAC (for cloning), and TBS7-Ptet-X-deGFP_Trps16,(for Ptet) were isolated from their cultures. After initial optimization runs, we prepared two different vectors at first. The vector to clone aln, pTAC, was digested with NdeI and HindII, having a fallout of about 500bp, after which the linearized band was cut and gel extraction was performed with it. The vector for Ptet was made by sequentially digesting pTAC plasmid with SphI and BglII and the linear band was cut and eluted, seeing a fall out of 200bp. After clone confirmation of Ptet, we immediately constructed the vector for cloning rhlAB as well, by digesting the clone with XbaI and SphI. Gel extraction of the linear band was performed and then eluted. All of the gels were run on 1% agarose.


Ligation, transformation and clone confirmation: For each cloning, respective inserts and vectors were ligated using 1:3 and 1:5 molar ratios and were transformed to freshly prepared E. coli DH5α cells. The colonies that came the next day were screened for colony PCR, using vector specific primers as well as gene specific primers. For alasan, we used Ptac_FP and aln_RP, for Ptet, we used gene specific forward and reverse primers. For rhlAB clones, we used Ptet_FP and rhlAB_RP. The colonies which came positive were selected and inoculated for plasmid isolation. For alasan, we immediately tested the colonies for Emulsification Index assay with a wide variety of oils. The colonies tested positive for EI assay, so we selected one of these to do our downstream work. The plasmids were then used for putting PCR against alasan gene, where all of them tested positive, thereby confirming our clone, Ptac-aln. The idea of cloning our gene under a tac promoter was that we could perform all our assays using the cloning strain, E. coli DH5α cells itself, they bypassing the need to use a specialized expression strain harboring a lysogenized phage expressing T7 RNA polymerase. The colony PCR images as well as the clone confirmation images are provided below with a detailed figure legend.



SDS-PAGE to detect the presence of the Aln protein: To visualize the expression of Aln protein in the heterologous host E. coli DH5𝛂, the cultures were induced with IPTG at the OD600 of 0.3-0.6. Samples were collected for 3 hours of induction from the cultures, and an empty vector containing just the promoter was used as a control. The cultures were sonicated to analyze the expression of the Alasan protein, the cell pellet was dissolved in 3 ml of sonication buffer and was sonicated using a 3 mm, sonication probe for 10 min with a pulse of 20 secs ON and 30 secs OFF cycle at an amplitude of 35%. The cultures were then centrifuged, the supernatant was collected. The pellet after sonication was also lysed. The samples were prepared by dissolving in laemmli dye containing SDS and 𝛃-mercaptoethanol, the samples were boiled at 95℃ for 10 min. These samples of both supernatant and pellet were run on 12% SDS-PAGE, to understand if the protein was soluble and can fold properly in the sonication buffer used. A protein of ~45kDa was observed in the supernatant of the induced sample harboring the clone with Aln gene. The same band was not visualized after staining and destaining in samples from pellet after sonication. This suggested that our protein was properly solubilized and was able to fold properly.


Assays to detect the type of biosurfactants: The induction of 1mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) activated the tac promoter under which aln gene has been cloned by us. As a result, we used 50ml induced culture after 3 hours of induction to extract alasan. Overnight lipid precipitation was done by conc. HCl, and then the next morning, we extracted the organic phase by using the solvent extraction method (chloroform: methanol=2:1). The upper aqueous layer was removed, and the lower layer was left to dry overnight. The honey coloured biosurfactant was then seen sticking to the walls of the flask. This was dissolved in deionized water and used for further studies. The working concentration of alasan we used was around 20 mg/ml.
Emulsification Index assay: Emulsification assay was carried out using petrol as it contains both aliphatic and aromatic hydrocarbons. Equal volume of 1:1 of petrol and cell free supernatant obtained after centrifugation of the overnight grown culture was added and mixed by vigorous vortexing for 2 min. The samples were kept for 24 h in a low vibrational area. The emulsification activity of each isolates was calculated by the given formula [1]:Emulsification index= (Height of the emulsion layer/Total height) ∗100


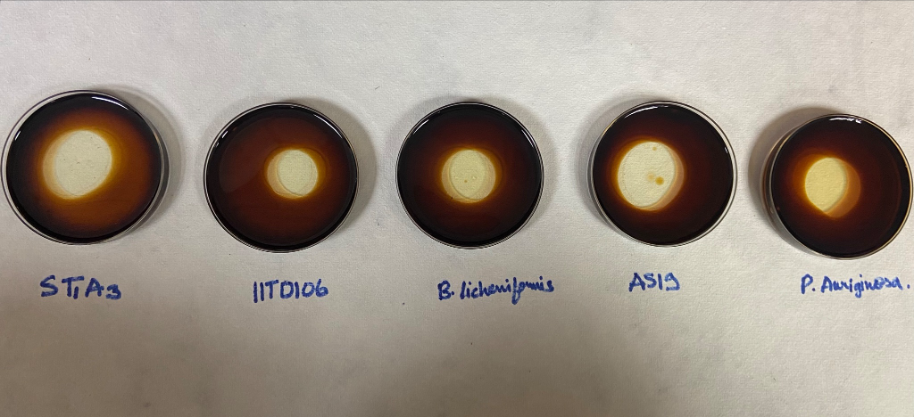
Oil displacement assay: We first used the crude biosurfactant for performing oil displacement assay with burnt motor oil (https://studio.youtube.com/video/4jjXcuqSytU/edit) ADD THIS VIDEO. We observed an instantaneous clearance zone caused by the recombinant alasan that we produced, thus leading us to infer it is functional (as was previously already noted after our positive EI assay).





FT-IR: for further confirmation of our TLC results, we decided to carry out IR spectroscopy with the dried sample for alasan. We detected peaks for primary and secondary amines and amides, conjugated alkenes C=C bonds, as well as N-H bond stretching, carboxylic acids. This further correlated to what we knew of the functional groups in the molecule. To the best of our knowledge, this is the first study of FT-IR conducted on alasan. The spectrum has been given below.

ICP-OES: In order to observe the extraction capacity of the biosurfactant on metals, ICP-OES assay was performed. The biosurfactant pellet weighing around 20 mg was dissolved in water with 100 ppm of salt solution. This sample was filtered to remove any contamination. It was then incubated for 12 hours at 200 rpm and 37 C, after which the sample was filtered again to remove the biosurfactant complexes so that only the metal left in the sample is detected. This was compared with the test 100 ppm salt solution which was treated through the same conditions. The assay was performed for three different metals, copper, chromium and cadmium. Alasan shows significant removal for all three metal ions. Copper was found to be reduced by 22%, Chromium 13.7% and Cadmium 8.42%. Following are the bar diagrams representing the same.

Contact Angle: Contact angle of the biosurfactant alasan was measured and found out to be around 33 degrees. The concentration of the sample was 170 mg/ml. Since it is much less than 90 degrees, it is likely to be a wet table and good biosurfactant.
