Register: Astrid, Dave, Emily, Hester, Lottie, Matt, Tobi, Yilu
Time: 10am-1pm
Today we delivered the equipment to the lab and started the briefing with Dr Markiv at 10am. This included safety briefing about bins, gloves and coats, as well as some core skills such as pipetting. Dr Markiv also gave us a quick tour of the facilities including the Faculty of Dentistry, Oral & Craniofacial Sciences. After going through our equipment, we decided what we might need to use from KCL's equipment and what our plan was for the rest of the week. We also found that in ordering the E.coli, they were not competent cells, as we had planned to use, so our timescales would have to accommodate the new protocols to prepare them for transformations.
Stock solutions prep for making competent E.coli:- 20g/L LB broth x2; 500ml water flask
- 0.1 M CaCl2 solution (7.35 g of CaCl2·2H2O in 500 ml of H2O) in a 500 ml Pyrex bottle. Autoclave and store at 4 oC.
- 0.1 M MgCl2 solution (10.165 g of MgCl2 ·6H2O in 500ml of H2O) in a 500ml Pyrex bottle. Autoclave and store at 4 oC.
- 50% glycerol solution (100 ml of glycerol and 100 ml H2O) in a 200 ml Pyrex bottle. Autoclave and store at 4 oC.
LB agar 2x 400ml in 500ml Pyrex bottle; 20g/L LB broth (8g) + 2% weight per volume agar (8g) in 400ml water
Close with sponge and foil. Label, Autoclave 125 oC/15 min and store at room temperature.
20g/L LB broth; 200ml
Close with sponge and foil. Label, Autoclave 125 oC/15 min and store at room temperature.
Ampicillin 100 mg/ml - 1 g in 10 ml of MilliQ water.
The stock solution is filtered through a 0.2 mm filter, aliquoted into 10 tips (1μ/ml each) and stored at -20oC until use. The final working concentration of all antibiotics is 1:1000 of the stock solution (1000x ampicillin).
Inoculating the E.coli cells onto agar plates which will have been prepared earlier.
Register: Astrid, Emily, Dave, Hester, Lottie, Tobi, Yilu
Time: 10am-1pm
Yesterday, we sorted our inoculation of our cells,the protocol of which can be found below, to make competent cells and transform them. We also found that the plasmids that we ordered, didn't have four random nucleotides on the ends of both restriction enzymes. As a result, they were too close to the end which made it more difficult for RNA polymerase to “break” the restriction enzymes..
- We used a mass spectrometer to measure the growth of E. Coli, based on its optical density in respect to the density of the media alone (in our case LB Broth)
- detects absorbance 600 Nm
- first measurement was 0.3, so we let the bacteria grow in a shaking incubator for 20 mins
- the second measurement was 0.585, which is within the range we needed (0.4-0.6)
- now we place cells on ice for 30 mins
- cells centrifuged in solutions
- leave on ice
- made agar plates
Primers:
- Need to order primers in order to amplify our GeneBricks as the restriction sites are too close to the end. For fwd Primer, take first 18 nucleotides, or slightly more as long as it ends on C or G. add 4 random nucleotides to the BEGINNING of the sequence
- for Rvs Primer, take the last 18 nucleotides and take the complementary bases, again ending in C or G. Add 4 nucleotides to the BEGINNING of the sequence. ACTG
- NOTE FOR FUTURE TEAMS: when ordering synthetic parts, always add 4 random nucleotides to the ends of your sequence so the restriction sites can be reached easily.
Inoculating and preparing cells:
K12 E.coli cells are being used (LZB 035) for transformations of both plasmids.- Work under flame.
- Streak 10 ml of E. coli cells onto one of the LB agar plates containing no antibiotic or specific for the cell type.
- Grow the cells overnight at 37 oC, no more than 16 h.
- Wrap plate in a parafilm and store at 4 oC.
Making competent cells:
- Pick a single colony of the cells from the LB agar plate into 10 ml of LB media containing no antibiotic or specific for the cell type. Grow the cultures overnight at 37 oC with shaking at 250 rpm.
- Inoculate 200 ml of prewarmed to 37oC LB medium (no antibiotics or specific for the cell type) with 10 ml of the overnight cultures, and grow at 37oC for 60 min, with vigorous shaking 250 rpm or until the OD600 is 0.4 - 0.5.
- Put the flask on ice for 30 min. At the same time chill sterile falcon (centrifuge) tubes.
- Aliquot culture into 50 ml each 4 x 50 ml chilled falcon (centrifuge) tubes.
- Harvest the cells by centrifugation for 7 min at 3500 rpm, at 4oC and discard supernatants completely.
- Resuspend cells in each tube in 12.5 ml of 0.1 M MgCl2.
- Centrifuge for 7 min at 3500 rpm, at 4oC and discard supernatants.
- Resuspend cells in each tube in 25 ml of 0.1 M CaCl2.
- Incubate cells on ice for 30 min.
- Centrifuge for 7 min at 3500 rpm, at 4oC and discard supernatants.
- Resuspend cells in each tube in 700 ml of 0.1 M CaCl2 and 300 ml of 50% glycerol. Final volume 1 ml in each tube.
- Aliquot 50 ml aliquots into 1.5 ml sterile microcentrifuge tube on ice and stored at -80oC.
Begin filming for our iGEM promotional video within the labs. + Order primers to add onto GeneBricks. + Turn GeneBricks into plasmids.
Register: Dave, Matt, Yilu
Time: 10am- 5pm
Beginning PCR for the THS T7 GFP and mCherry circuits, then confirming amplification with gel electrophoresis.
5ng/mL - PCR Prep 25microL GoTaq Master Mix 2x concentrated (using 2-20 pipette, 10 and 15) 10 microL from template DNA (either 1 - mCherry and 2 - GFP) 5microL of each primers (6F and 6R) 5microL of nuclease free water Start with larger volume first, so that there is no liquid left in the pipette when inserting into pipette Using centrifuge to get rid of air bubbles in the tubes A-test 95 52 - annealing temperature 72 1% agarose x2
- Stir the gel and TAE buffer with avoiding getting the sediment up the side
- >1kb of DNA use 1% agarose, and \<1kb DNA use <1% agarose
- Microwave until the gel powder dissolves into the buffer, without over microwaving (which would lower the concentration)
1kb DNA ladder 10microL GelRed Nucleic acid into each 1% gel agarose Pour into the tray with comb 100W power
Register: Dave, Matt, Yilu
Time: 2pm-5pm
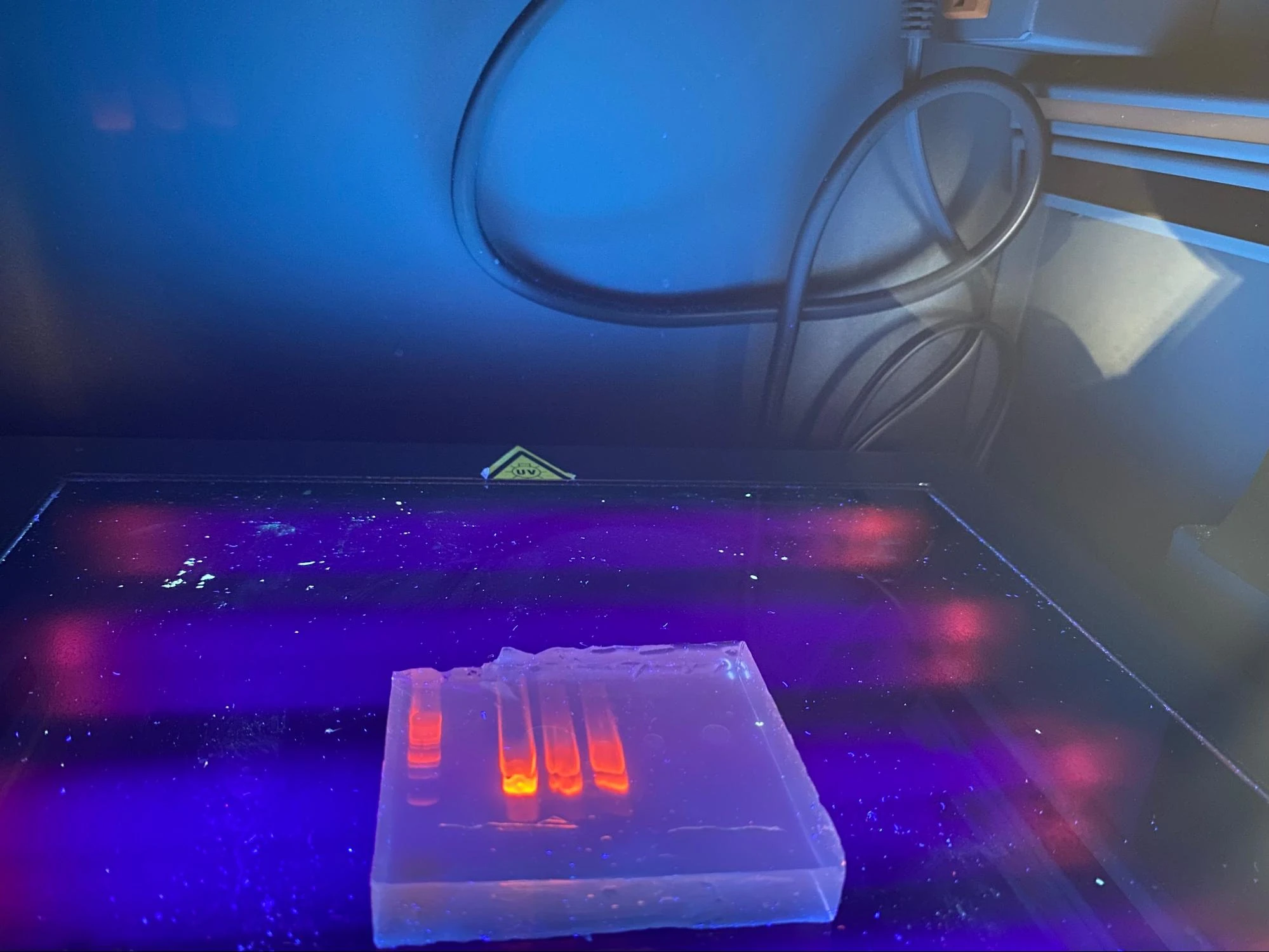
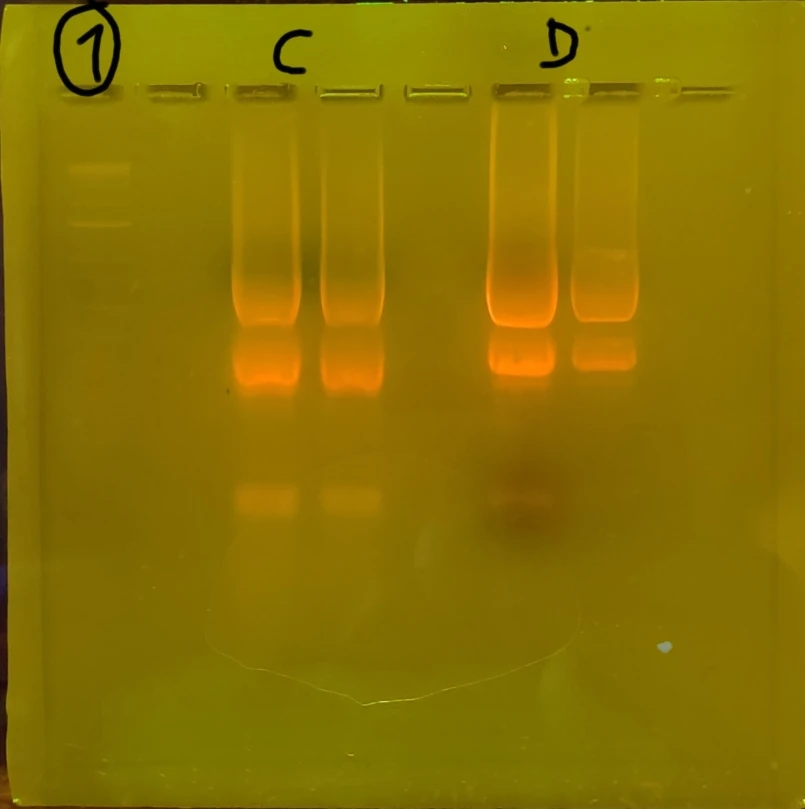
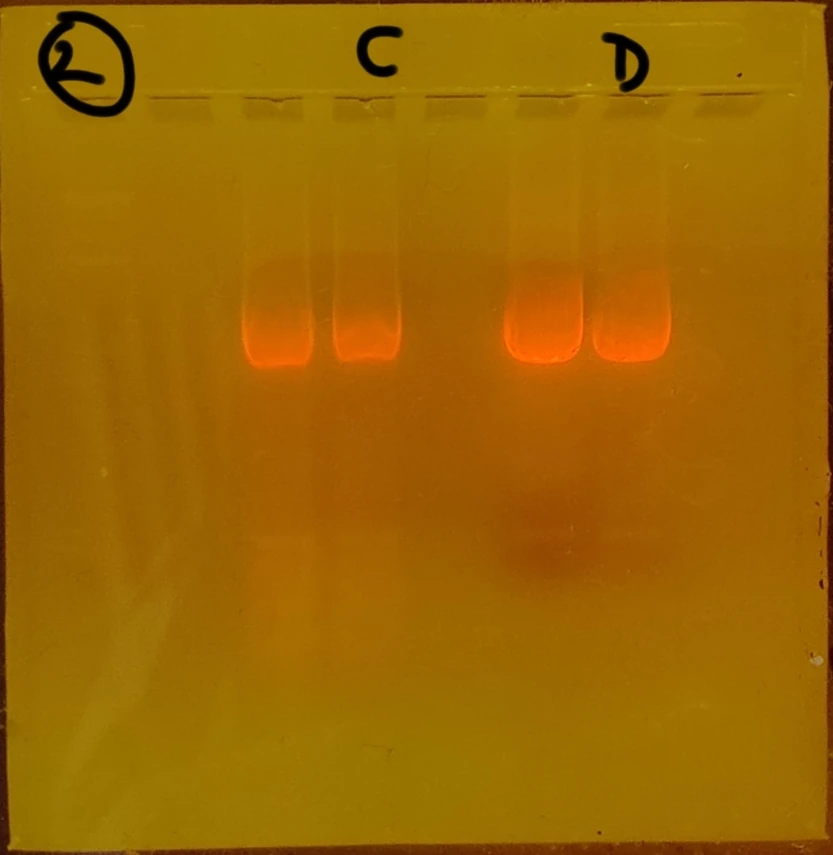
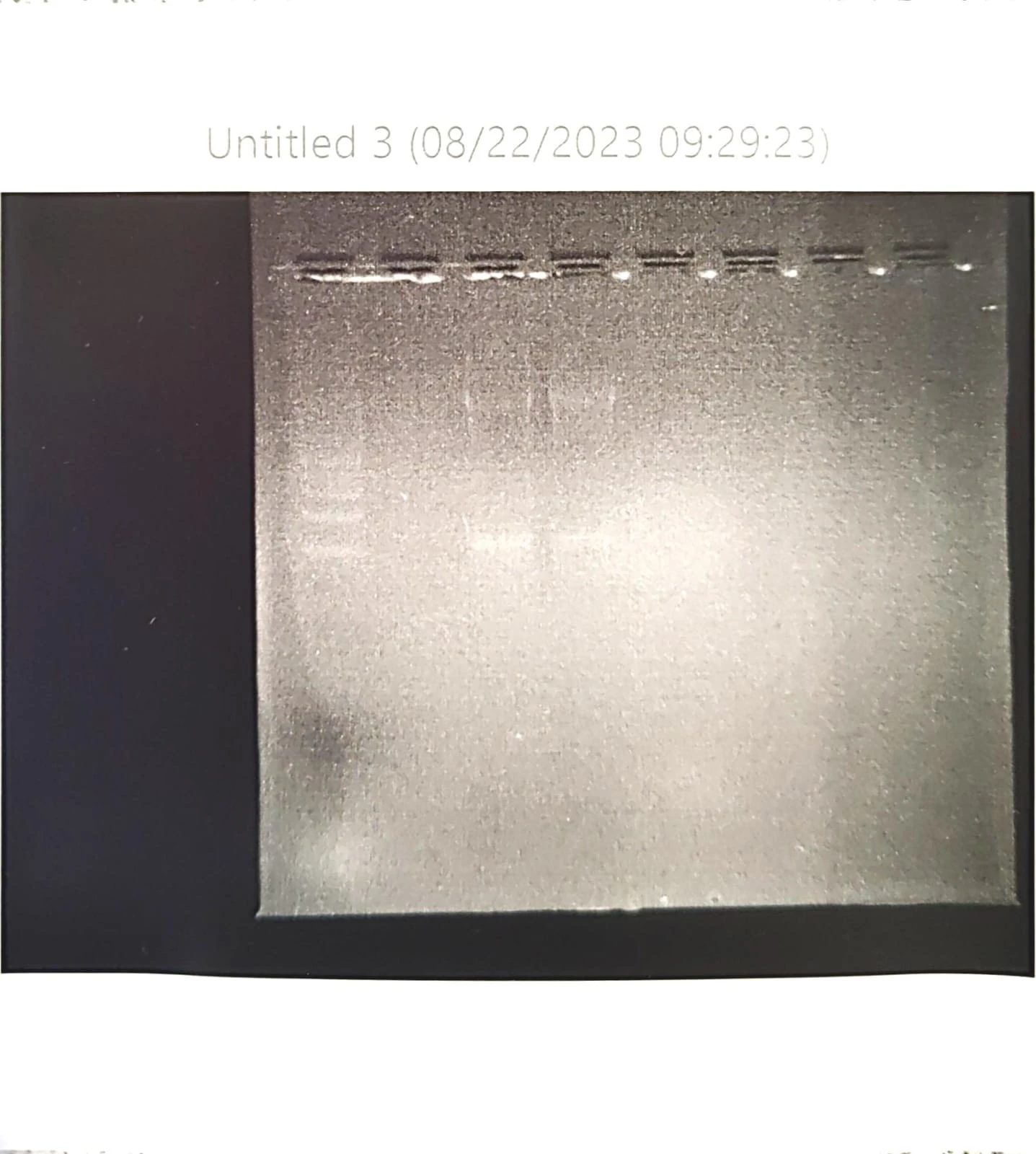
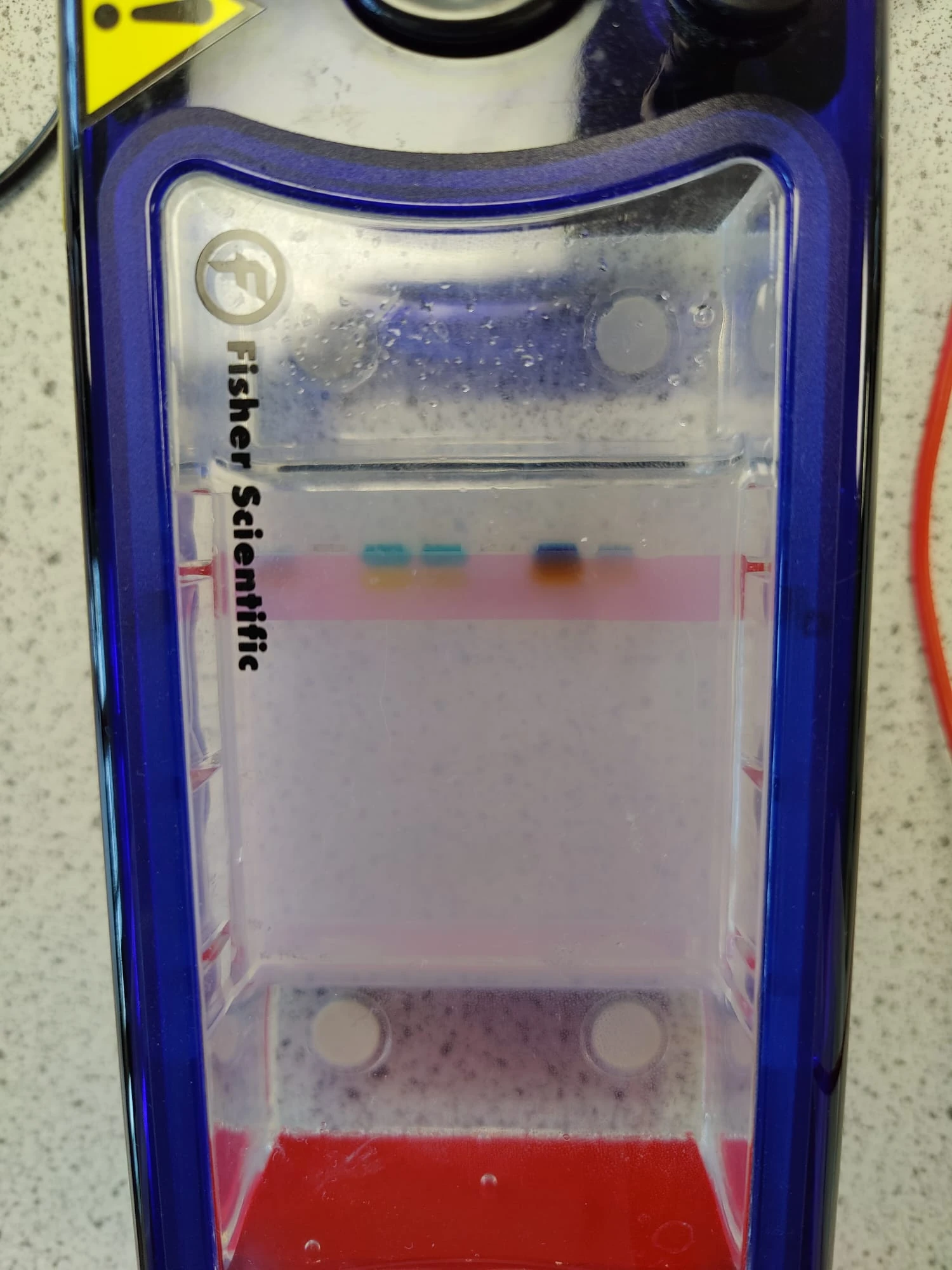
Repeated PCR prep with different annealing temperature A-test 95 55 72 Changed agarose gel to 2% 10microL GelRed Nucleic acid Took photos not under UV to prevent mutation risk of the DNA. Used a blue light filter box. The second set (GFP) of gel electrophoresis was successful, showing only one band which is what we expected to be shown from the amplification / PCR and one lower down for the primers (fewer base pairs). The first set however showed 3 bands. The faintest being primers again, but the larger ones harder to "diagnose". Through comparison between the first and second gels, as well as the sample ladder, it seemed most likely that the top band was the sequence that we wanted. We removed both regardless.
Transform linear DNA into plasmids using backbone; Insert our plasmids into competent e. Coli; test our THS's
Register: Matt, Hester, Yilu
Time: 11am-3pm
Repeated PCR again - difficulty with the number of bands shown in gel electrophoresis Possibly due to problems with volumes of primers used (unequal) Found that tap water was used, instead of distilled water, which explains why the number of bands did not correlate with the results which we expected, as there were contaminants which would have been removed in dH2O (as a precipitated solid). 10 microL of stock primer and 90 microL of water (FWD and REV) New master mix (perhaps a different level of ATCG-ase to help with the annealing) GoTaq Long PCR Master Mix 2x A-test thermocycle temperature 95 - denaturing temperature 57 - annealing temperature (increased from yesterday) 72 - extension temperature 20 mL TAE buffer 50x, fill to 1L pure water 2.4g of agarose, 120mL of TAE buffer dilute 10microL Blue/Orange 6X Loading Dye to GoTaq Long PCR Master Mix tube (=60microL) Load the agarose gel with 30:20 GoTaq (Green) and 30:30 GoTaq Long Difficulties with the actual toehold switch mechanism: regarding E.coli vs human ribosomal activity and whether proteins will be expressed
Register: Hester, Lottie, Yilu
Time: Unknown
- What exactly do we want to do?
- Two fluorescent proteins and many miRNA?
- Which backbone do we want to use to clone the plasmid DNA
- Organise a call with Dr Markiv to discuss our new plans
- Choose a backbone
- ampicillin or chloramphenicol - ask him to tell us which antibiotic to use
- Check the validity of our TXTL protocol
- Organise wet lab availability with all members of the team to continue lab work later in the year.