Biology
Project description
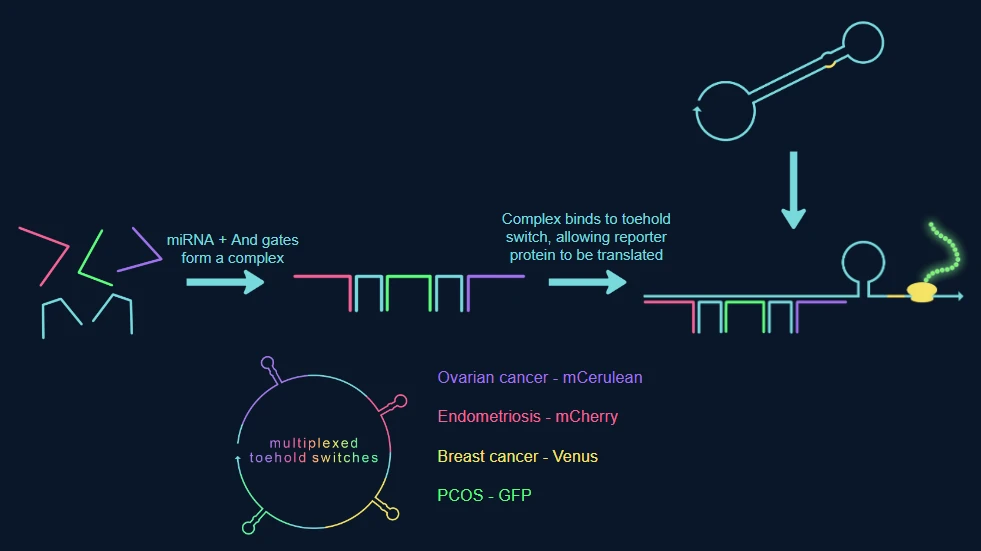
The project this year has focused on combating the antiquated tests currently in place for diseases linked to women’s health. Our method involves multiplexed regulation which uses four stacked toehold switches, for the four diseases, which each test for three miRNA. The switches incorporate genetic 'AND' gates, allowing for high specificity. Uniquely, our project allows for the miRNA binding sites to be swapped meaning various diseases can be tested for, using the collaborative database that we have started (you can find this on our Contribution page).The presence of the miRNA can be tested simultaneously using one blood sample, with the turnaround time for results being significantly improved compared to older tests. The toehold switches being used will synthesise different fluorescent proteins, once they have unfolded, given the presence of the correct miRNA. The chosen fluorescent proteins have varying wavelengths so can be tested separately, as shown on our Hardware page.
To increase the accessibility of our test, we have chosen to use miRPA to amplify the miRNA taken in our sample. This allows the miRNA to be isothermally amplified, and therefore doesn’t require the use of an expensive thermocycler. More information on the probes used can be found on our Software page.
Our Three-Step Process
Step 1: Extraction of miRNA
Extraction of miRNA is done using electromagnetic particles.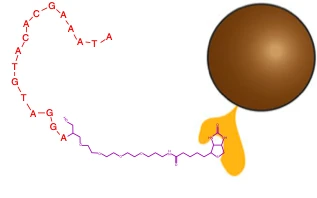
These are the steps: Samples of blood are taken from a patient and added to a solution containing magnetic nanoparticles. These nanoparticles often consist of a metal (usually iron) oxide and are also coated with a biotin-streptavidin bonded anti-miRNA strand. A set of electromagnets is turned on and off in quick succession, causing the nanoparticles to move and stir the solution. During this process, target miRNAs bond to the complementary anti-miRNA via base-pairing. Afterwards, the magnetic beads are pulled to the side of the container while the rest of the solution is removed. The miRNA is removed from the beads using an elution buffer. The previous steps are repeated three times in order to remove the maximum possible amount of miRNA for amplification.
Step 2: miRPA
As seen in RIBOTOX, it is difficult to discriminate between leaky expression of toehold switches and <2.25M concentrations of miRNAs, and there is no data giving concentrations of the trigger miRNAs we are concerned with online.Therefore, to ensure miRNA concentration is high enough for them to be detected by the toehold switches, we need to amplify our microRNAs. Not only is PCR difficult to perform on microRNAs due to their short length, we need to find a solution that will allow amplification to take place isothermally, in a single tube, increasing the accessibility of our tests.
Recombinase Polymerase Amplification (RPA) is a single tube, isothermal alternative to PCR, which can amplify dsDNA strands. So, before RPA can work, we need to reverse transcribe miRNA into DNA. This can be done using miRPA.
Two DNA probes, one with 5’ phosphorylation, bind to the miRNA, and are ligated together by DNA ligase. Then, primers are added, with DNA polymerase, and complementary strands to the ligated probes are synthesised. Then, RPA can take place: primers, which are associated with recombinase protein dislodge the strands, replicating them in a similar method to PCR, but as no heat cycles are required to break up the strands, the process can take place isothermally.
In order for the miRNA to be detected, we use ‘asymmetric RPA’: an excess of forward primers are added (usually 5x the amount), so an excess of the strand that was originally miRNA form is produced, so there is now ssDNA with the miRNA code in DNA. This can be detected by toehold switches. In order to design probes for miRPA, we can use NUPACK’s design functions in its API to find probes which can bind to the miRNAs, but have overhangs which do not bind within themselves, to ensure primers can easily anneal to them.
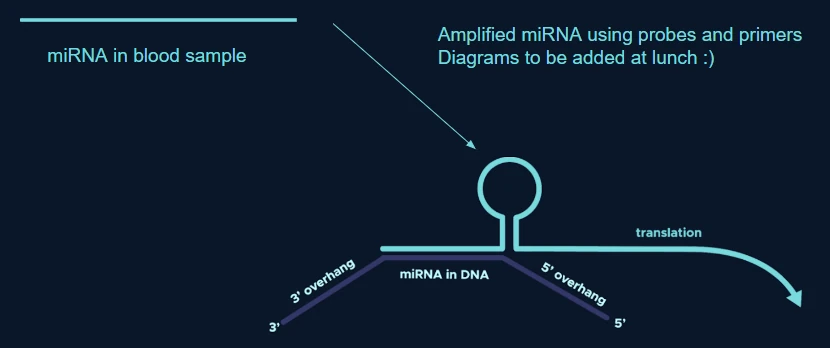
Step 3: Charaterisation of toehold switches
To detect this now amplified miRNA, we made use of toehold switches. Conventionally, toehold switches open in response to the presence of a specific trigger, usually a single miRNA strand, however this can create issues regarding specificity due to how many miRNA several different conditions can share. Our solution is to use AND gates made of RNA to detect a combination of miRNA all specific to 1 disease, solving this specificity problem.
The RNA and gates are specially designed to join miRNA strands for a single disease together and create a trigger complex, the switches are designed to have a binding site that is complementary to the trigger complex’ unpaired bases. When the trigger complex binds to the switch, the switch collapses, exposing the RBS and start codon allowing for the translation of a reporter protein.
As seen earlier, our switches are multiplexed allowing us to detect 4 diseases at once. Each switch has their own binding site and reporter protein depending on the condition the switch is designed to detect.
MicroRNA
Our project uses microRNA biomarkers to indicate the presence of disease. MicroRNAs (miRNAs) are small, highly conserved non-coding RNA molecules involved in the regulation of gene expression.[1]These microRNAs circulate in the blood, and can be shown to be either upregulated, or downregulated in certain conditions. The dysregulation of these diseases can be linked to developmental stages of diseases[2] and by using toehold switches, which have binding sites for the upregulated miRNA we can detect the disease. While our system is meant to be modular, we have chosen endometriosis, PCOS, ovarian cancer and breast cancer to prove the theory. Given we needed 3 miRNA for each disease we have chosen the following, which are all upregulated:
Breast Cancer:
miRNA-24-3p: can be used for early detection [3]
miRNA-21-3p: has different expression patterns in serum of breast cancer [4]
miRNA-373-5p [5]
PCOS:
Ovarian Cancer:
Endometriosis:
Toehold Switches
Toehold switches are synthetic RNAs which are able to use RNA fragments to bring about transcription of certain proteins, mimicking the role of a messenger RNA from within a normal cell. In its OFF state, the toehold switch remains in a hairpin structure with an 'input' recognition site for the RNA, and a hidden or inaccessible ribosome binding site. This allows for the synthesis of proteins under exact biological conditions and therefore allows for the detection of specific or complex environments. [14] Within Genoswitch, we use microRNA as a biomarker to indicate when a specific blood environment, brought about by a certain disease, occurs. [15] When the miRNA is sufficiently upregulated, the ribosome binding site for the various fluorescent proteins is unfolded, decreasing the length of the stem and allowing for exposure of the start codon, thus allowing for translation.miRPA
As described by project RIBOTOX, it is difficult to discriminate between leaky expression of toehold switches and <2.25M concentrations of miRNAs. [16] We therefore needed to amplify the miRNA. We have chosen to use miRPA, similarly to the RIBOTOX project. It is a single tube, isothermal alternative to PCR so both increases accessibility to groups with less access to thermocyclers and also is easier to perform amplification on miRNA than PCR.We therefore needed to amplify the miRNA. We have chosen to use miRPA, similarly to the RIBOTOX project. It is a single tube, isothermal alternative to PCR so both increases accessibility to groups with less access to thermocyclers and also is easier to perform amplification on miRNA than PCR.
Two DNA probes, one with 5' phosphorylation, bind to the miRNA, and are ligated together by DNA ligase. Then, primers are added with DNA polymerase, and complementary strands to the ligated probes are synthesised. Then, RPA can take place: primers, associated with recombinase protein dislodge the strands, replicating them in a similar method to PCR but as no heat cycles are required to break up the strands, the process can take place isothermally.
In order for the miRNA to be detected, we will need to use 'asymmetric RPA': an excess of forward primers are added (usually 5x the amount), so an excess of the strand that was originally miRNA form, so there is now ssDNA with the miRNA code in DNA. This can be detected by toehold switches. In order to design probes for miRPA, we can use NUPACK's design functions in its API to find probes which can bind to the miRNAs, but have overhangs which do not bind within themselves, to ensure primers can easily anneal to them.
References
1. ^ MacFarlane, L., and Murphy, P, R., "MicroRNA: Biogenesis, Function and Role in Cancer", Curr Genomics., 11(7): 537-561, 2010, Available: https://doi.org/10.2174/138920210793175895
2. ^ Wang, J., Chen, J., and Sen, S., "microRNA as Biomarkers and Diagnostics.", Journal of Cellular Physiology, 231(1): 25-30, 2022, Available: https://doi.org/10.1002/jcp.25056
3. ^ An, X., Quan, H., Lv, J., Meng, L., Wang C., Yu, Z., et al., "Serum microRNA as potential biomarker to detect breast atypical hyperplasia and early-stage breast cancer.", Future Oncology, 14(30): 3145-3161, Dec. 2018, Available: https://doi.org/10.2217/fon-2018-0334
4. ^ Mar-Aguilar, F., Mendoza-Ramírez, J, A., Malagón-Santiago, I., Espino-Silva, P, K., Santuario-Facio, S, K., Ruiz-Flores, P., Rodríguez-Padilla, C., and Reséndex-Pérez, D., "Serum Circulating microRNA Profiling for Identification of Potential Breast Cancer Biomarkers", Disease Markers, 34(3): 163-169, Mar. 2013, Available: https://doi.org/10.3233/DMA-120957
5. ^ Müller, V., et al, "Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the Geparquinto trial", Breast Cancer Research and Treatment, 147(1), 61-68, Aug. 2014, Available: https://doi.org/10.1007/s10549-014-3079-3
6. ^ Butler, A, E., et al., "microRNA Expression in Women With and Without Polycystic Ovarian Syndrome Matched for Body Mass Index", Frontiers in Endocrinology, 11, Apr. 2020, Available: https://doi.org/10.3389/fendo.2020.00206
7. ^ Ham, X, M., Tian, P, Y., Zhang, J, L.,, "MicroRNA-486-5p inhibits ovarian granulosa cell proliferation and participates in the development of PCOS via targeting MST4", European Review for Medical and Pharmacological Sciences, 23, 7217-7223, Sept. 2019, Available: https://doi.org/10.26355/eurrev_201909_18823
8. ^ Motahari Rad, H., et al., "Characterization of altered microRNAs related to different phenotypes of polycystic ovarian syndrome (PCOS) in serum, follicular fluid, and cumulus cells", Taiwanese Journal of Obstetrics and Gynecology, 61(5), 768–779, Sept. 2022, Available: https://doi.org/10.1016/j.tjog.2022.05.013
9. ^ Zhang, L., Chen, H., He, F.-X., Zhang, S., Li, A., Zhang, A., & Zhang, A., "MicroRNA-320a Promotes Epithelial Ovarian Cancer Cell Proliferation and Invasion by Targeting RASSF8", Frontiers in Endocrinology, 11, Feb. 2021, Available: https://doi.org/10.3389/fonc.2021.581932
10. ^ Priscila, Margiotti, K., Fabiani, M., Mateus Camargo Barros-Filho, Sparacino, D., Cima, A., Longo, S., Cupellaro, M., Mesoraca, A., & Giorlandino, C., "Multi-analytical test based on serum miRNAs and proteins quantification for ovarian cancer early detection", PLOS ONE, 16(8), e0255804-e0255804, Aug. 2021, Available: https://doi.org/10.1371/journal.pone.0255804
11. ^ Xiao, Y., Bi, M., Guo, H., & Li, M., "Multi-omics approaches for biomarker discovery in early ovarian cancer diagnosis", eBioMedicine, 79, 104001–104001, Apr. 2022, Available: https://doi.org/10.1016/j.ebiom.2022.104001
12. ^ Kolanska, K., Bendifallah, S., Canlorbe, G., Mekinian, A., Touboul, C., Aractingi, S., Chabbert-Buffet, N., Daraï, E., "Role of miRNAs in Normal Endometrium and in Endometrial Disorders: Comprehensive Review", Journal of Clinical Medicine, 10(16), 3457-3457, Aug, 2021, Available: https://doi.org/10.3390/jcm10163457
13. ^ Monnaka, V.U., Hernandes, C., Heller, D., Podgaec, S., "Overview of miRNAs for the non-invasive diagnosis of endometriosis: evidence, challenges and strategies. A systematic review", einstein (São Paulo), 19, eRW5704, Sept, 2020, Available: https://doi.org/10.31744/einstein_journal/2021RW5704
14. ^ Wyss Institute, "Toehold Switches for Synthetic Biology", , Aug, 2018, Available: https://wyss.harvard.edu/technology/toehold-switches-for-synthetic-biology
15. ^ Wang S, Emery NJ, Liu AP, "A Novel Synthetic Toehold Switch for MicroRNA Detection in Mammalian Cells", ACS Synthetic Biology, 8(5), 1079-1088, Apr. 2019, Available: https://doi.org/10.1021/acssynbio.8b00530
16. ^ City of London iGEM Team 2021, "Design", RIBOTOX, 2021, Available: https://2021.igem.org/Team:City_of_London_UK/Design